Detecting cell-type-specific AEI in bulk RNA-seq
Methods Overview
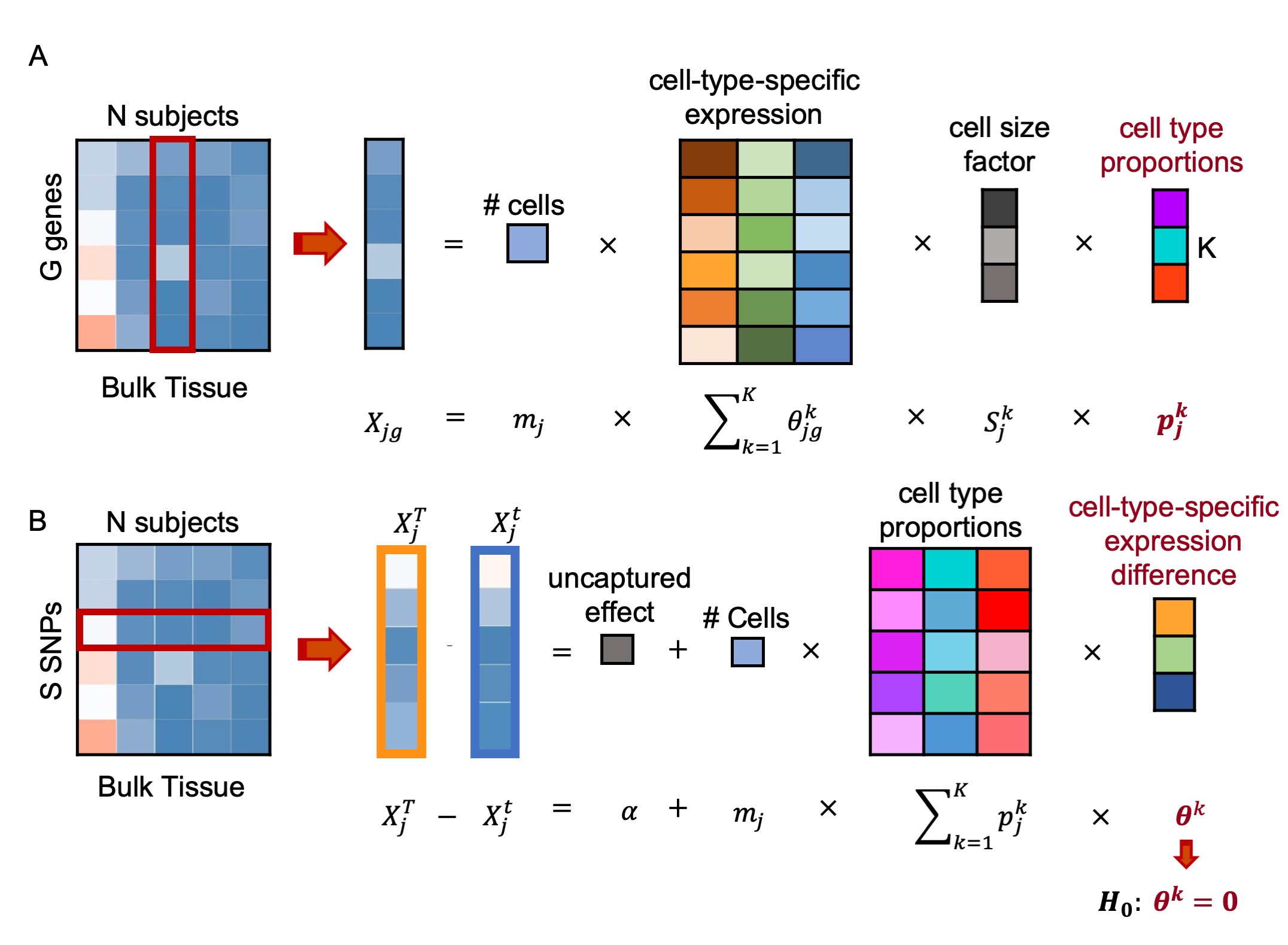
We proposed BSCET, a two-step regression-based procedure that allows us to integrate ulk and ingle-cell RNA-seq data to detect ll-ype-specific allelic expression imbalance (AEI). In the first step, BSCET performs cell-type deconvolution analysis to infer cell type composition in bulk RNA-seq data using scRNA-seq data as the reference. In the second step, given estimated cell type proportions of bulk RNA-seq samples, BSCET tests cell-type-specific AEI by aggregating allele-specific bulk RNA-seq read counts across individuals. Since the degree of AEI may vary with disease phenotypes, we further extend BSCET to incorporate clinical factors such as disease status to infer covariate effects on cell-type-specific AEI.
How to cite BSCET
Please cite the following publication:
Detecting cell-type-specific allelic expression imbalance by integrative analysis of bulk and single-cell RNA sequencing data
J. Fan, X. Wang, R. Xiao, M. Li
PLOS Genetics 17(3): e1009080. https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1009080
Developed by Jiaxin Fan (jiaxinf@pennmedicine.upenn.edu).